 |
Protein sequence comparisons typically double the evolutionary look-back time over DNA sequence comparisons.
|
 | The requirement for a common folded structure in homologous proteins usually causes these proteins to be similar over the entire length of the gene product (or domain). Therefore, most sequences that share statistically significant similarity throughout their entire lengths are homologous.
|
 | Matches that are more than 50% identical in a 20-40 amino acid region occur frequently by chance.
|
 | Distantly related homologs may lack significant similarity. Two or more homologous sequences may have very few absolutely conserved residues.
|
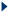 | If homology has been inferred due to significant similarity scores between two proteins, A and B, that align over their entire lengths and between protein B and a third protein, C, then proteins A and C must also be homologous, even if they share no significant similarity.
|
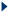 | Low complexity regions, transmembrane regions and coiled-coil regions frequently display significant similarity in the absense of homology. Low complexity regions can be filtered out using the default parameters of BLAST. Transmembrane and coiled-coil regions should be identified and masked (by eliminating these regions from the query) by the user.
|
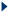 | Results of searches using different scoring systems may be compared directly using normalized scores. If S is the (raw) score for a local alignment, the normalized score S' (in bits) is calculated by the formula S'=(lambdaS-lnK)/ln2. lambda and K are parameters associated with a given scoring system.
|
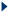 | A normalized score, S' with E value = E, is statistically significant if it exceeds log N/E where N is the size of the search space.
|
 | As the evolutionary distance between two sequences increases, the length of a local alignment required to achieve a statistically significant score also increases.
|

