Alignment Scoring
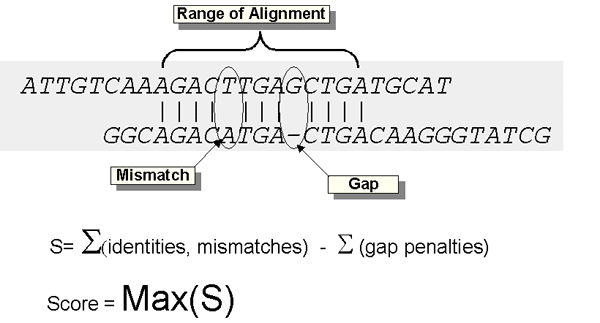
Calculating alignment scores. The raw score S for an alignment is calculated by summing the scores for each aligned position and the scores for gaps. In this figure, a DNA alignment is shown. In amino acid alignments, the score for an identity or a substitution is given by the specified substitution matrix (e.g. BLOSUM62). BLAST 2.0 and PSI-BLAST use "affine gap costs" which charge the score -a for the existence of a gap, and the score -b for each residue in the gap. A gap of k residues therefore receives a total score of -(a+bk) and a gap of length 1 receives the score -(a+b). Gap creation and extension variables a and b are inherent to the scoring system in use (BLAST 2.0 defaults).